Introduction
This post provides a step-by-step guide to visualizing changes in global life expectancy using the data from the World Bank group through its package wbstats and the ggplot2 library in R. The post demonstrates various techniques for creating an informative and visually appealing line plot, including arranging the order of faceted panels, creating efficient visualizations of summary statistics, displaying graphic elements that extend beyond the plot boundary, making unique annotations in selected faceted panels, and loading custom fonts.
Packages and data cleaup
Getting data from the World Bank
It is very simple to use wbstats if you know the names of the indicators from the World Development Indicators database. Today we download the 3 indicators used in gapminder: life expectancy, GDP per capita, and the total population. We also get the database of countries that we can look at.
iso3c date iso2c country gdp_capita life_expectancy pop
1 ABW 1960 AW Aruba NA 64.152 54608
2 ABW 1961 AW Aruba NA 64.537 55811
3 ABW 1962 AW Aruba NA 64.752 56682
16663 ZWE 2020 ZW Zimbabwe 1372.697 61.124 15669666
16664 ZWE 2021 ZW Zimbabwe 1773.920 59.253 15993524
16665 ZWE 2022 ZW Zimbabwe 1676.821 59.391 16320537The current dataset consists of individual countries around the world, lacking information about their respective continents. In order to link each country with its corresponding continent, we require an additional file containing a mapping of countries to continents.
By combining this supplementary file with our original dataset using country names as the main identifier, we can enhance our data with continental details. This process is essential for conducting continent-level analysis and developing visual representations that offer a more comprehensive geographical perspective.
theme_set(theme_classic(base_size = 12))
f1 = lf.data |>
ggplot(aes(x = year, y = life_expectancy,
color = continent, fill = continent )) +
geom_line(aes(group = country), alpha = .2)+
stat_summary(fun = mean, geom = "line", size = 2)+
scale_x_continuous(breaks = seq(1960,2020,20))+
scale_y_continuous(limits = c(30, 85))
f1
f2 = f1+
facet_wrap(~continent, nrow = 1)+
ggsci::scale_color_aaas()+
ggsci::scale_fill_aaas() +
theme(legend.position = "none")
f2 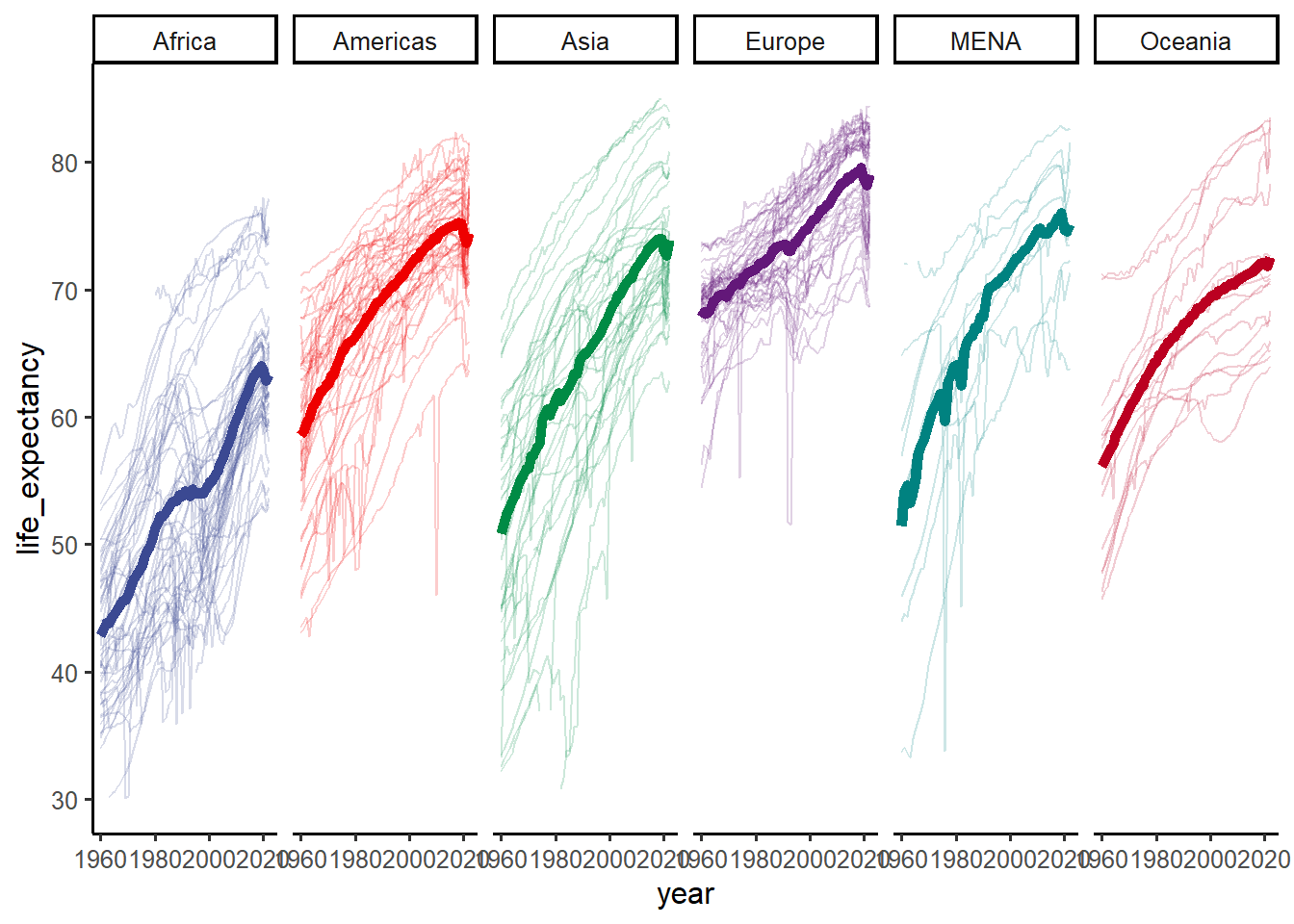
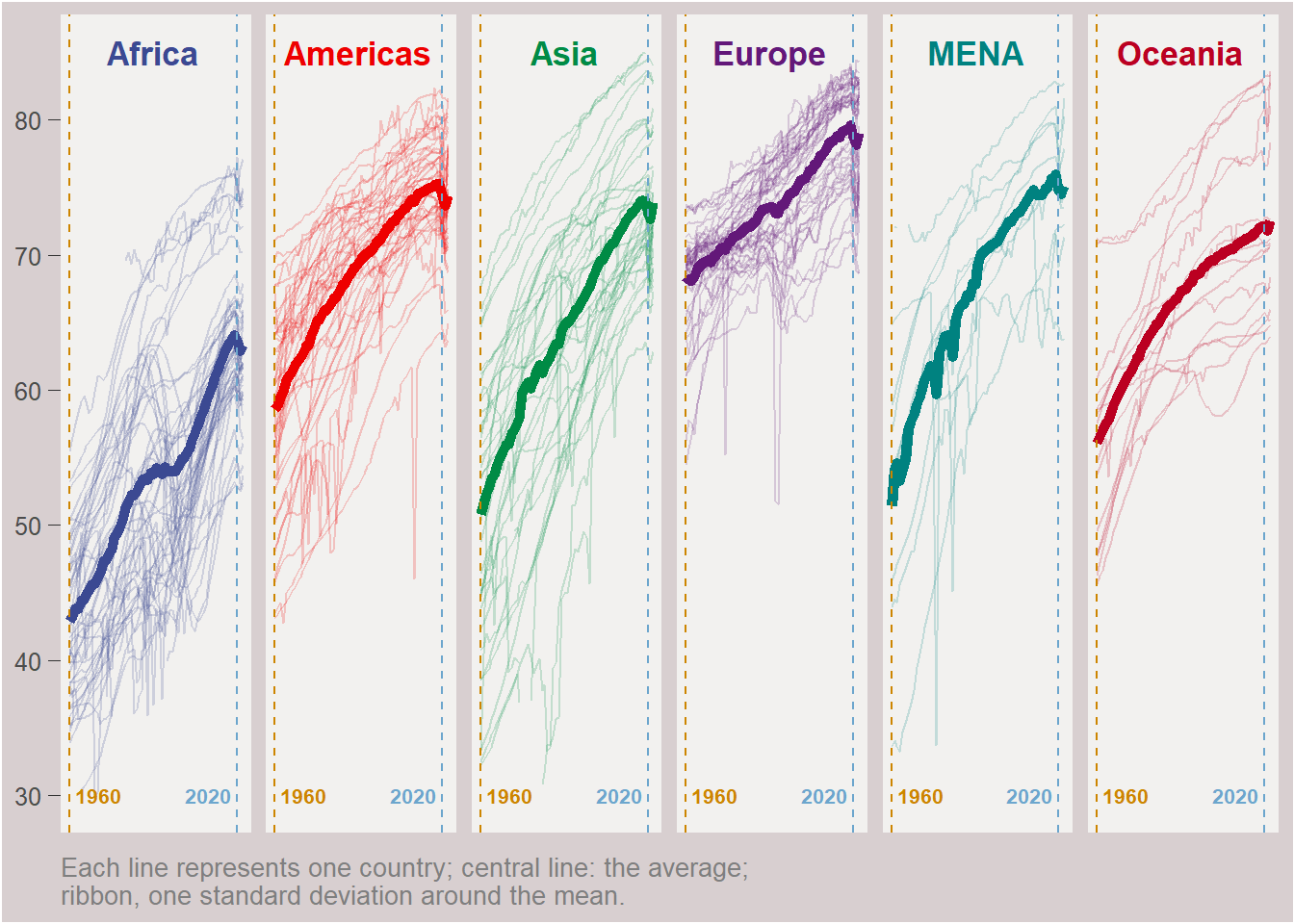
f3 = f2 +
# year 1960
geom_vline(xintercept = 1960, linetype = "dashed", color = "orange3") +
# year 2020
geom_vline(xintercept = 2020, linetype = "dashed", color = "skyblue3") +
# add text annotation
# year 1952
annotate(geom = "text", x = 1960, y = 30, label = " 1960",
fontface = "bold", size = 2.8, hjust = 0, color = "orange3") +
# year 2007
annotate(geom = "text", x = 2020, y = 30, label = "2020 ",
fontface = "bold", size = 2.8, hjust = 1, color = "skyblue3")
f3
life.1960_2020 <- lf.data %>%
filter(year %in% c(1960, 2020)) %>%
group_by(continent, year) %>%
summarise(life_expectancy = mean(life_expectancy, na.rm = T) %>% round())
life.1960_2020 |> head()# A tibble: 6 × 3
# Groups: continent [3]
continent year life_expectancy
<chr> <int> <dbl>
1 Africa 1960 43
2 Africa 2020 64
3 Americas 1960 59
4 Americas 2020 74
5 Asia 1960 51
6 Asia 2020 74panel.titles <- lf.data |> distinct(continent) |> arrange(continent)
f3 +
# not clip graphical elements beyond the panel range
coord_cartesian(clip = "off") +
geom_text(
data = panel.titles,
aes(x = 1980, y = 85, label = continent),
size = 4.5, nudge_x = 10, fontface = "bold") +
# titles
labs(
# title = "Steady increase of Human Life Expectancy",
caption = "Each line represents one country; central line: the average; \nribbon, one standard deviation around the mean.",
x = NULL) +
theme(
strip.text = element_blank(),
axis.line.y = element_blank(),
axis.line.x = element_blank(),
axis.text.x = element_blank(),
axis.ticks.x = element_blank(),
axis.title.y = element_blank(),
axis.ticks.length.y = unit(5, "pt"),
axis.ticks.y = element_line(linetype = "solid", linewidth = .15),
plot.title = element_text(size = 18, family = "fat"),
plot.caption = element_text(hjust = 0, size = 10, color = "grey50"),
plot.background = element_rect(fill = "#d8cfd0"),
panel.background = element_rect(fill = "#f2f1ef")
) 
Conclusion
This post describes the process of creating an annotated line plot in R using the ggplot2 library to visualize the changing capture fish landings, with a focus on the Kenya, Uganda and Tanzania.
Citation
@online{semba2024,
author = {Semba, Masumbuko},
title = {Create Lineplots Using Ggplot2 to Visualize Changes in Global
Life Expectancy},
date = {2024-06-02},
url = {https://lugoga.github.io/kitaa/posts/visualize_line2/},
langid = {en}
}