Introduction
Bio-ORACLE is a freely available global dataset that provides a comprehensive set of bioclimatic variables — physical, chemical, biological and topographic data layers with global extent and uniform resolution for modelling the distribution of marine biodiversity. One of the challenges in working with large environmental datasets like Bio-ORACLE is managing and integrating the data into your modeling workflows. Fortunately, Samuel Bosch and Salvador Fernandez -Bosch and Fernandez (2023) developed sdmpredictors package in R language that simplify access of wide range of environmental data.
The sdmpredictors package (Bosch and Fernandez, 2023) provides several key functions to simplify access and use of the Bio-ORACLE data layers:
Listing Available Layers: The package includes a function to list all the available data layers in the Bio-ORACLE dataset, making it easy to browse and select the variables you need for your analysis.
Extracting Data: With just a few lines of code, you can extract the specific data layers you want to use, subsetting by geographic region or resolution as needed.
Managing Data: The package handles the conversion and management of the data, ensuring it’s in a format that’s ready to be integrated into your bioclimatic modeling pipelines.
Integration with Modeling Workflows: sdmpredictors seamlessly integrates with popular species distribution modeling packages like dismo and MaxEnt (Hijmans et al., 2023), simplifying the process of incorporating the Bio-ORACLE data into your modeling workflows.
Let’s begin by loading packages that we are going to use in this session as highlighted in the chunk below;
List available Bio-ORACLE layers
With the list_datasets function, you can view all the available datasets. If you only want terrestrial datasets then you have to set the marine parameter to FALSE and vice versa.
By default, all datasets are returned. If marine, freshwater, and terrestrial are all set to FALSE, only datasets without land or sea masks are included.
List layers
Using the list_layers function we can view all layer information based on datasets, terrestrial (TRUE/FALSE), marine (TRUE/FALSE) and/or whether it should be monthly data.
By default, it returns all layers from all datasets. If both marine and terrestrial are set to FALSE, only layers from datasets without land or sea masks are returned. You can list layers for paleo and future climatic conditions using the functions list_layers_paleo and list_layers_future. To find available paleo and future climate layers for a given current climate layer code, use the functions get_paleo_layers and get_future_layers.
Load layers
The load_layers function is used to specifically load the layers that you want to work with. This function provides a way for you to select and access the specific layers within your data that are relevant to your analysis. By using the load_layers function, you can focus on the specific components of your data that are important for your particular research or analysis, helping you to streamline your workflow and avoid unnecessary processing of irrelevant information.
robin = "+proj=robin +lon_0=0 +x_0=0 +y_0=0 +ellps=WGS84 +datum=WGS84 +units=m +no_defs"
ggplot() +
geom_spatraster(data = tmax.bottom)+
geom_sf(data = spData::world)+
scale_fill_gradientn(name = "Temperature", colours = oce::oce.colors9A(120), trans = scales::modulus_trans(p = 0.2), na.value = NA)+
labs(title = "Maximum temperature at the sea bottom (ºC)")+
coord_sf(crs = robin)+
cowplot::theme_map()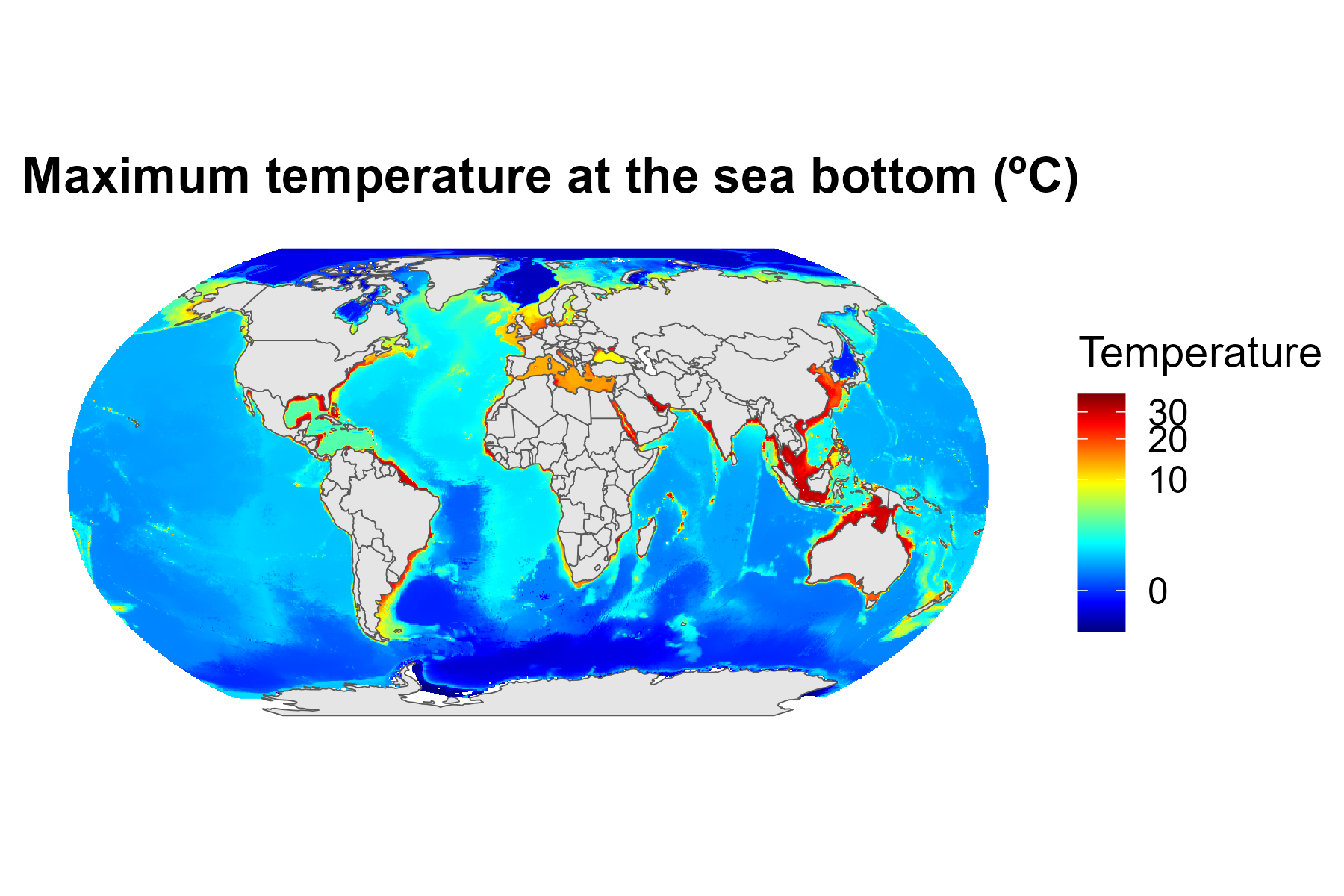
Loading future data
The list_layers_future function gives detailed information about future climate layers in one or more datasets, including metadata like layer names, descriptions, time periods covered, and specific variables or conditions represented. This code chunk is used to analyze future marine layers in climate datasets. The list_layers_future function is used to retrieve information about future climate layers, specifically filtering out terrestrial layers to focus on marine and freshwater layers. This allows to examine future climate data related to marine environments.
Then, the unique(future$scenario) function is called to retrieve and display the unique scenarios available in the future climate layers. Scenarios represent different projections or models of future climate conditions based on various assumptions and inputs. By identifying the unique scenarios, users can better understand the range of future climate conditions represented in the datasets and select the appropriate data for their analysis.
Loading paleo data
To explore paleo climate data, we start by using the list_layers_paleo function. By setting the parameter terrestrial = FALSE, we specifically filter out terrestrial layers, focusing on marine and possibly freshwater layers. This function retrieves detailed information about the available paleo climate layers.
After loading the paleo data, we can identify the unique epochs represented in the datasets using the unique(paleo$epoch) function. This helps in understanding the different historical periods covered by the data.
We can also identify the unique models used to generate the paleo climate data by using the unique(paleo$model_name) function. This helps to understand the different modeling approaches and sources of the paleo climate data.
Additional function
To retrieve detailed information about specific layers, we can use the get_layers_info function. In this example, we are interested in the layers “BO_calcite,” “BO_B1_2100_sstmax,” and “MS_bathy_21kya.” The function call returns common information about these layers.
To obtain the equivalent future layer codes for specific current climate layers, you can use the get_future_layers function. This function helps to find the future projections of the given current climate layers based on specified scenarios and years. In this example, we are looking for the future layer codes for “BO_sstmax” and “BO_salinity” under the “B1” scenario for the year 2100.
# looking up statistics and correlations for marine annual layers
datasets <- list_datasets(terrestrial = FALSE, marine = TRUE) |> list_layers()
# layers <- list_layers(datasets)
# filter out monthly layers
layers <- layers[is.na(layers$month),]
layer_stats(layers)[1:2,]
correlations <- layers_correlation(layers)Summarize
This post provides a basic overview to get started with sdmpredictors package in R for exploring and using environmental data in species distribution modeling. Adjust the functions and parameters as per your specific research needs and dataset requirements.
References
Citation
@online{semba2024,
author = {Semba, Masumbuko},
title = {Simplifying {Access} to {Bio-ORACLE} {Data} with the
Sdmpredictors {R} {Package}},
date = {2024-06-25},
url = {https://lugoga.github.io/kitaa/posts/bio_oracle/},
langid = {en}
}